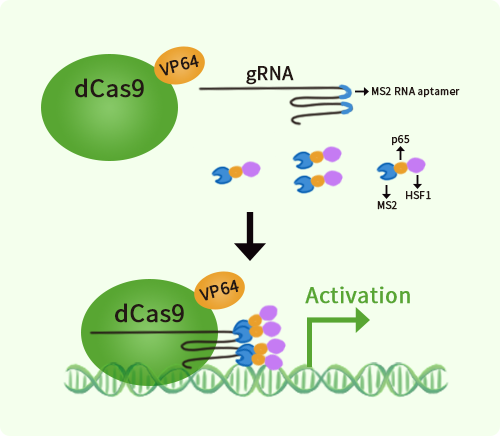
Human Claudin 3 (CLDN3) activation kit by CRISPRa
CAT#: GA100955
CLDN3 CRISPRa kit - CRISPR gene activation of human claudin 3
Find the corresponding CRISPRi Inhibitor Kit
USD 1,290.00
2 Weeks*
Specifications
| Product Data | |
| Format | 3gRNAs, 1 scramble ctrl and 1 enhancer vector |
| Symbol | CLDN3 |
| Locus ID | 1365 |
| Kit Components | GA100955G1, Claudin 3 gRNA vector 1 in pCas-Guide-GFP-CRISPRa GA100955G2, Claudin 3 gRNA vector 2 in pCas-Guide-GFP-CRISPRa GA100955G3, Claudin 3 gRNA vector 3 in pCas-Guide-GFP-CRISPRa 1 CRISPRa-Enhancer vector, SKU GE100056 1 CRISPRa scramble vector, SKU GE100077 |
| Disclaimer | The kit is designed based on the best knowledge of CRISPa SAM technology. The efficiency of the activation can be affected by many factors, including nucleosome occupancy status, chromatin structure and the gene expression level of the target, etc. |
| Reference Data | |
| RefSeq | NM_001306 |
| Synonyms | C7orf1; CPE-R2; CPETR2; HRVP1; RVP1 |
| Summary | Tight junctions represent one mode of cell-to-cell adhesion in epithelial or endothelial cell sheets, forming continuous seals around cells and serving as a physical barrier to prevent solutes and water from passing freely through the paracellular space. These junctions are comprised of sets of continuous networking strands in the outwardly facing cytoplasmic leaflet, with complementary grooves in the inwardly facing extracytoplasmic leaflet. The protein encoded by this intronless gene, a member of the claudin family, is an integral membrane protein and a component of tight junction strands. It is also a low-affinity receptor for Clostridium perfringens enterotoxin, and shares aa sequence similarity with a putative apoptosis-related protein found in rat. [provided by RefSeq, Jul 2008] |
Documents
| Product Manuals |
| FAQs |
Resources
Other Versions
| SKU | Description | Size | Price |
|---|---|---|---|
| KN404882 | CLDN3 - KN2.0, Human gene knockout kit via CRISPR, non-homology mediated. |
USD 1,290.00 |
{0} Product Review(s)
Be the first one to submit a review
Product Citations
Kraguljac, White, Hadley et al
Schizophr Bull (2016) 42 (4), 1046-55 DOI: 10.1093/schbul/sbv228
Soneson, Fontes, Zhou et al
Neurobiol Dis (2010) 40 (3), 531-43 DOI: 10.1016/j.nbd.2010.07.013
Yan, Zhou, Ying et al
Comput Med Imaging Graph (2013) 37 (4), 272-80 DOI: 10.1016/j.compmedimag.2013.04.007
Smittenaar, Guitart-Masip, Lutti et al
J Neurosci (2013) 33 (46), 18087-97 DOI: 10.1523/JNEUROSCI.2167-13.2013
Haas, Mills, Yam et al
J Neurosci (2009) 29 (4), 1132-9 DOI: 10.1523/JNEUROSCI.5324-08.2009






























































































































































































































































 Germany
Germany
 Japan
Japan
 United Kingdom
United Kingdom
 China
China