Ikaros (IKZF1) Human Gene Knockout Kit (CRISPR)
CAT#: KN413207
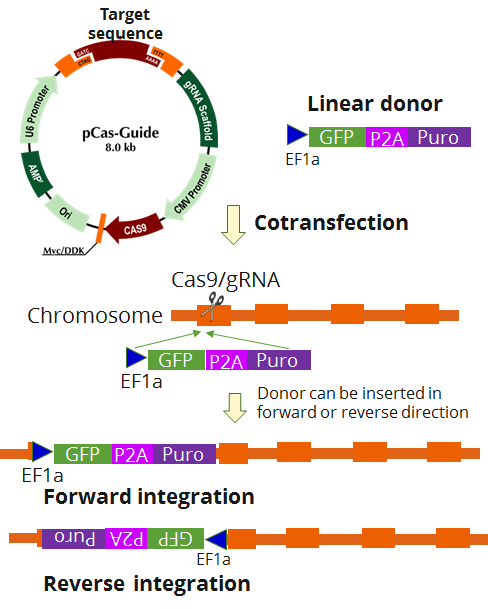
IKZF1 - KN2.0, Human gene knockout kit via CRISPR, non-homology mediated.
KN2.0 knockout kit validation
KN413207 is the updated version of KN213207.
USD 1,290.00
2 Weeks*
Size
Other products for "IKZF1"
Specifications
| Product Data | |
| Format | 2 gRNA vectors, 1 linear donor |
| Donor DNA | EF1a-GFP-P2A-Puro |
| Symbol | IKZF1 |
| Locus ID | 10320 |
| Disclaimer | The kit is designed based on the best knowledge of CRISPR technology. The system has been functionally validated for knocking-in the cassette downstream the native promoter. The efficiency of the knock-out varies due to the nature of the biology and the complexity of the experimental process. |
| Reference Data | |
| RefSeq | NM_001220765, NM_001220766, NM_001220767, NM_001220768, NM_001220769, NM_001220770, NM_001220771, NM_001220772, NM_001220773, NM_001220774, NM_001220775, NM_001220776, NM_001291837, NM_001291838, NM_001291839, NM_001291840, NM_001291841, NM_001291842, NM_001291843, NM_001291844, NM_001291845, NM_001291846, NM_001291847, NM_006060 |
| Synonyms | Hs.54452; IK1; IKAROS; LyF-1; LYF1; PPP1R92; PRO0758; ZNFN1A1 |
| Summary | This gene encodes a transcription factor that belongs to the family of zinc-finger DNA-binding proteins associated with chromatin remodeling. The expression of this protein is restricted to the fetal and adult hemo-lymphopoietic system, and it functions as a regulator of lymphocyte differentiation. Several alternatively spliced transcript variants encoding different isoforms have been described for this gene. Most isoforms share a common C-terminal domain, which contains two zinc finger motifs that are required for hetero- or homo-dimerization, and for interactions with other proteins. The isoforms, however, differ in the number of N-terminal zinc finger motifs that bind DNA and in nuclear localization signal presence, resulting in members with and without DNA-binding properties. Only a few isoforms contain the requisite three or more N-terminal zinc motifs that confer high affinity binding to a specific core DNA sequence element in the promoters of target genes. The non-DNA-binding isoforms are largely found in the cytoplasm, and are thought to function as dominant-negative factors. Overexpression of some dominant-negative isoforms have been associated with B-cell malignancies, such as acute lymphoblastic leukemia (ALL). [provided by RefSeq, May 2014] |
Documents
| Product Manuals |
| FAQs |
| SDS |
Resources
Other Versions
| SKU | Description | Size | Price |
|---|---|---|---|
| GA107033 | IKZF1 CRISPRa kit - CRISPR gene activation of human IKAROS family zinc finger 1 |
USD 1,290.00 |
{0} Product Review(s)
0 Product Review(s)
Submit review
Be the first one to submit a review
Product Citations
*Delivery time may vary from web posted schedule. Occasional delays may occur due to unforeseen
complexities in the preparation of your product. International customers may expect an additional 1-2 weeks
in shipping.






























































































































































































































































 Germany
Germany
 Japan
Japan
 United Kingdom
United Kingdom
 China
China