NABC1 (BCAS1) Human Gene Knockout Kit (CRISPR)
CAT#: KN423329
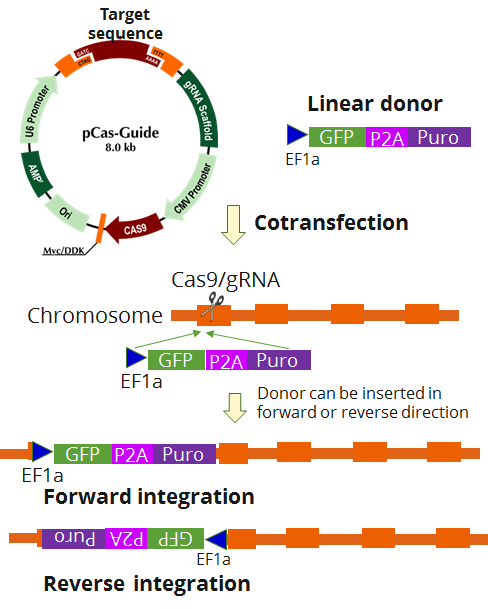
BCAS1 - KN2.0, Human gene knockout kit via CRISPR, non-homology mediated.
KN2.0 knockout kit validation
KN423329 is the updated version of KN223329.
USD 1,290.00
2 Weeks*
Size
Other products for "BCAS1"
Specifications
| Product Data | |
| Format | 2 gRNA vectors, 1 linear donor |
| Donor DNA | EF1a-GFP-P2A-Puro |
| Symbol | BCAS1 |
| Locus ID | 8537 |
| Disclaimer | The kit is designed based on the best knowledge of CRISPR technology. The system has been functionally validated for knocking-in the cassette downstream the native promoter. The efficiency of the knock-out varies due to the nature of the biology and the complexity of the experimental process. |
| Reference Data | |
| RefSeq | NM_001316361, NM_001323347, NM_003657, NM_001366295, NM_001366296, NM_001366297, NM_001366298 |
| Synonyms | AIBC1; NABC1 |
| Summary | This gene resides in a region at 20q13 which is amplified in a variety of tumor types and associated with more aggressive tumor phenotypes. Among the genes identified from this region, it was found to be highly expressed in three amplified breast cancer cell lines and in one breast tumor without amplification at 20q13.2. However, this gene is not in the common region of maximal amplification and its expression was not detected in the breast cancer cell line MCF7, in which this region is highly amplified. Although not consistently expressed, this gene is a candidate oncogene. [provided by RefSeq, Apr 2016] |
Documents
| Product Manuals |
| FAQs |
| SDS |
Resources
Other Versions
| SKU | Description | Size | Price |
|---|---|---|---|
| GA105651 | BCAS1 CRISPRa kit - CRISPR gene activation of human breast carcinoma amplified sequence 1 |
USD 1,290.00 |
{0} Product Review(s)
0 Product Review(s)
Submit review
Be the first one to submit a review
Product Citations
*Delivery time may vary from web posted schedule. Occasional delays may occur due to unforeseen
complexities in the preparation of your product. International customers may expect an additional 1-2 weeks
in shipping.






























































































































































































































































 Germany
Germany
 Japan
Japan
 United Kingdom
United Kingdom
 China
China